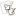P. ramorum species-specific primer pair
Nested within the Phytophthora genus-specific amplicon (cox spacer region)
- Alignments of cox spacer genus-specific amplicon used in primer development ( .msf, .nex, fastA)
- Primers FMPr-1a + FMPr-7
- FMPr-1a (dGTATTTAAAATCATAGGTGTAATTTG)
- FMPr-7 (dTGGTTTTTTTAATTTATATTATCAATG)
- amplicon is 135 bp in size (sequence for genus-specific amplicon)
- Good amplification at 2, 3 or 4 mM Mg, weaker at 1 mM
- Does not amplify well at 65 C, does work at 64 C annealing
Species specificity for this primer pair for other Phytophthora spp. was tested on samples of Phytophthora DNA that had a first round amplification with Phy-8b+10b. When tested on 10 ng total DNA there was some faint background bands produced for P. erythroseptica, P. palmivora, P. citrophthora, and P. drechsleri (but a different size than the diagnostic fragment).
- Using a nested amplification procedure with purified P. ramorum DNA the limit of detection was 2 fg total DNA
- Adding plant DNA extracted with the Fast DNA kit of Qbiogene reduced the sensitivity of detection
- Table of Phytophthora species tested and isolate history
![]()
![]()
A) Test for species specificity of amplification with the P. ramorum species-specific primer pair (FMPr-1a + FMPr-7) with a 1:100 dilution of the first round amplification with the Phytophthora genus-specific primers FMPh-8b + FMPh-10b observed in Figure 2 as the target DNA. The annealing temperature was 64° C. B) Amplification of different isolates of P. ramorum from California and Europe with the P. ramorum species-specific primer pair (FMPr-1a + FMPr-7). The template DNA was a 1:100 dilution of products of the first round amplification with the Phytophthora genus-specific primers FMPh-8b + FMPh-10b using an initial concentration of 20 ng of target DNA for each isolate. A total of 12 µl of the amplification mixture was loaded into each well of a 3% NuSieve 3:1 agarose gel. Isolate Prg-2 T is the type culture for this species. The molecular size marker is a 100-bp ladder from New England BioLabs.
In addition to the Phytophthora spp. noted above the P. ramorum species-specific primers did not amplify bands when FMPh-8b + FMPh-10b amplicons from the following species were used as a template DNA: P. clandestine, P. humicola, P. ideai, P. iranica, P. katsurae, P. medii, P. melonis, P. inflata, P. primulae, P. richardiae, P. porri, P. quercina, P. tentaculata, or isolates from three of the tentative species groupings described by Brasier et al. (2003)(P. taxon Raspberry, P. taxon Pgchlamydo, or Phytophthora sp. “O” group).
Test for amplification of plant DNA
![]()
Limit of detection
The sensitivity of the detection system was evaluated by using a dilution series of purified DNA from P. ramorum that had been spectrophotometrically quantified for amplification with the genus-specific primer pair. This was then diluted 1:25 and 1 µl was added to a second round amplification with the nested P. ramorum species-specific primer pair. To evaluate the influence of plant DNA extracted with the Fast Prep procedure by the CDFA noted below on the sensitivity of detection, parallel samples of the P. ramorum dilution series were spiked with 2 µl of DNA from healthy plants prior to amplification with the genus-specific primer pair.
A faint genus-specific band was observed with total DNA concentrations of 200 fg for isolate Prg-2 of P. ramorum, below which no amplified products were observed. When this was diluted 1:25 and 1 µl used as target DNA for amplification with the P. ramorum specific primer pair, the limit of detection was 2.0 fg. Similar results were obtained for two other isolates with the exception that one of the isolates had an amplified species-specific band at 20.0 but not 2.0 fg. Spiking the DNA dilution series with 2 µl plant DNA extracted at the CDFA (the same rate that would be used for evaluation of field samples) reduced the level of sensitivity by different amounts depending on the plant species. For U. californica the sensitivity was reduced by a factor of approximately 100 (from 2 fg to 200 fg) while for A. macrophylum, A. menziesii and R. californica the sensitivity was reduced by a factor of approximately 1000 with less amplification observed when spiking with A. macrophylum DNA than the other two plant species.
These results highlight the need for optimization of extraction methods for isolating DNA from infected plant material to ensure the full sensitivity of the marker system can be obtained. Clearly, the extraction method used in this current investigation needs to be further optimized to take advantage of the level of sensitivity of the described mitochondrial marker system.
![]()
Limit of detection of P. ramorum using a dilution series of total DNA. A) Amplification with the Phytophthora genus-specific primers followed by nested amplification with the P. ramorum species-specific primer pair using 1 µl of a 1:25 dilution of the first round amplification with the genus-specific primer pair. B) Nested amplification of purified P. ramorum DNA with the species-specific primer pair when the first round amplification with the genus-specific primer pair was spiked with water or DNA from four different plant species. A total of 12 µl of the amplification mixture was loaded into each well of a 3% NuSieve 3:1 agarose gel. The molecular size marker is a 100-bp ladder from New England BioLabs.
Sensitivity compared to the ITS markers
The sensitivity of the described mitochondrial marker system compared to the ITS marker system approved by USDA-APHIS was evaluated by using a dilution series of purified DNA from P. ramorum that had been spectrophotometrically quantified for amplification with the respective first round primers of each marker system. This was then diluted 1:25 and 1 µl was added to a second round amplification with the nested primer pair.
The sensitivity of nested amplifications for the two marker systems are comparable, however, the ITS marker is more robust with just a single round of amplification. One possible reason for this could be the high level of stringency used with the first round amplification for the mitochondrial markers to prevent background amplification of plant or Pythium species. Reducing the stringency (eliminating the glycerol and/or reducing the annealing temperature) enhances the sensitivity of the first round of amplification and hence, the level of detection of the second round nested amplification. However, if the Phytophthora genus-specific amplicon alone is used as a diagnostic marker for pathogen detection this would be feasible only if the plant material used in the investigation did not cause background amplification.
Mitochondrial marker system
![]()
Limit of detection of P. ramorum using a dilution series of total DNA from three different isolates of the pathogen. A) Amplification with the Phytophthora genus-specific primers FMPh-8b + FMPh-10b with 2% glycerol in the reaction mixture and an annealing temperature of 66° C. B) Amplification with the P. ramorum species-specific primer pair (FMPr-1a + FMPr-7) using 1 µl of a 1:25 dilution of the first round amplification with the genus-specific primer pair from the above gel picture. In both pictures a total of 12 µl of the amplification mixture was loaded into each well of a 3% NuSieve 3:1 agarose gel. The molecular size marker is a 100-bp ladder from New England BioLabs.
ITS Marker system
![]()
Multiplex amplification
Addition of the Phytophthora genus-specific primer pair at a concentration of 0.1 µM to this second round amplification enhanced the sensitivity of detection at the genus level in cases where the target Phytophthora DNA was present in low concentrations; however, higher primer concentrations reduced amplification of the P. ramorum diagnostic fragment. Combining both the Phytophthora genus-specific and P. ramorum species-specific primers in a second round multiplex amplification produced an amplification profile that was different from using the primers individually due to generation of additional amplicons between these primer pairs that are 293 to 296 bp in size.
![]()
Amplified products of Phytophthora ramorum using A) the Phytophthora genus-specific primer pair (FMPh-8b + FMPh-10b), B) the P. ramorum species-specific primer pair (FMPr-1a + FMPr-7) and C) multiplex amplification containing 0.1 µM final concentrations of the Phytophthora genus-specific primer pair (FMPhy-8b + FMPhy-10b) and 1.0 µM final concentrations of the P. ramorum species-specific primer pair (FMPr-1a + FMPr-7). A total of 12 µl of the amplification mixture was loaded into each well of a 3% NuSieve 3:1 agarose gel.
Amplification from symptomatic plant tissue
DNA extracted from symptomatic field tissue was provided by Cheryl Blomquist of the CDFA. Extractions were done with a FastDNA kit from Qbiogene in accordance with the manufacturers instructions. Amplifications were carried out using the parameters described above.
The results obtained with the mitochondrial marker system correlated well with the results obtained at the CDFA for pathogen recovery from symptomatic tissue and with PCR amplification for P. ramorum, using the rDNA ITS P. ramorum-specific primers of Garbelotto et al. (2002). For P. ramorum no differences in results were obtained; all samples that scored positive for P. ramorum in the CDFA laboratory also were positive with the mitochondrial markers. The results for four of these samples were validated by sequence analysis of the Phytophthora genus-specific amplicon and comparison to data from purified cultures of this pathogen. The mitochondrial marker system also identified 16 additional samples infected with other Phytophthora spp. (P. nemorosa, P. pseudosyringae, and P. syringae) that could not be identified with the ITS marker system. With a few explainable exceptions, these results correlated well with pathogen recovery in the CDFA laboratory. One exception was a sample from a Manzanita sp. from which a P. ilicis-like isolate was cultured. Due to the small size of the lesions the tissue samples for DNA extraction were different from the samples plated on selective medium; the lack of amplification of the Phytophthora genus-specific amplicon in this sample (907B below) could be due to another pathogen causing the lesion that was used for DNA extraction. Another exception was amplification of a Phytophthora genus-specific amplicon in DNA extracted from symptomatic U. californica tissue from which a Phytophthora sp. was not cultured. Sequence analysis of the Phytophthora genus-specific amplicons confirmed the band was not an artifact and that they were identical to P. nemorosa. Recovery of these pathogens from symptomatic tissue can be inconsistent, with greater success in the wetter winter and spring months and a lower frequency of recovery during the drier summer and fall months (observations of C. Blomquist of CDFA and J. Davidson & other members of D. Rizzo’s lab). These plant samples were collected on June 10, which may have contributed to the lack of pathogen isolation.
![]()
A) First round multiplex amplification of field samples with primer pairs FMPl-2b + FMPl-3b and FMPh-8b + FMPh-10b using 2 µl of DNA from the FastPrep extraction. A total of 12 µl of the amplification mixture added to each well of a 1.5% agarose gel. The molecular size marker is a 100 bp ladder from New England Biolabs. B) Second round multiplex amplification of a 1:25 dilution of the first multiplex amplification using primer pairs FMPh-8b + FMPh-10b (0.1 µM) and FMPr-1a + FMPr-7 (1.0 µM). A total of 12 µl of the amplification mixture was loaded into each well of a 3% NuSieve 3:1 agarose gel. The molecular size marker is a 100-bp ladder from New England BioLabs.
|